Create test matrix(造数据)
set.seed(6)test = matrix(rnorm(200), 20, 10)test[1:10, seq(1, 10, 2)] = test[1:10, seq(1, 10, 2)] + 3test[11:20, seq(2, 10, 2)] = test[11:20, seq(2, 10, 2)] + 2test[15:20, seq(2, 10, 2)] = test[15:20, seq(2, 10, 2)] + 4colnames(test) = paste("Test", 1:10, sep = "")rownames(test) = paste("Gene", 1:20, sep = "")正文从这里开始
# Show text within cellspheatmap(test, display_numbers = TRUE)pheatmap(test, display_numbers = TRUE, number_format = "\\%.1e")pheatmap(test, display_numbers = TRUE, display_numbers = matrix(ifelse(test > 5, "*", ""), nrow(test)))display_numbers = T 即Show text within cells
number_format 可以格式输出display_number
或者干脆自定义一个matrix通过display_numbers参数进行display
# legend(图例)的设置选项pheatmap(test) # p1pheatmap(test, legend_breaks = -1:7) # p2pheatmap(test, legend_breaks = 1:6, legend_labels = c("6","6", "6", "6", "6", "6")) # p3pheatmap(test, legend_breaks = 9:14, legend_labels = c("6","6", "6", "6", "6", "6")) # p4pheatmap函数会在内部算出legend的数值范围,本例中大概是 -1:7
在数值范围内,我们可以设定legend_breaks,以及对legend_breaks这个label的文本展示
legend_breaks和legend_labels是有一个对应关系的,否则报错如下
Error in pheatmap(test, legend_breaks = 1:6, legend_labels =
c(“6”, “6”, : Lengths of legend_breaks and legend_labels must be
the same假如我们的设定范围超出,就如p4所示

# Fix cell sizes and save to file with correct sizepheatmap(test, cellwidth = 15, cellheight = 12, main = "Example heatmap") # the title of the plotpheatmap(test, cellwidth = 15, cellheight = 12, fontsize = 8, filename = "test.pdf") # save to pdf# Change angle of text in the columns (0, 45, 90, 270 and 315)pheatmap(test, angle_col = "45")pheatmap(test, angle_col = "0")有关annotation
# Generate annotations for rows and columns(先造数据)annotation_col = data.frame( CellType = factor(rep(c("CT1", "CT2"), 5)), Time = 1:5 )rownames(annotation_col) = paste("Test", 1:10, sep = "")annotation_row = data.frame( GeneClass = factor(rep(c("Path1", "Path2", "Path3"), c(10, 4, 6))) )rownames(annotation_row) = paste("Gene", 1:20, sep = "")# Display row and color annotationspheatmap(test, annotation_col = annotation_col, cluster_cols = F)pheatmap(test, annotation_col = annotation_col, annotation_legend = FALSE, cluster_cols = F)pheatmap(test, annotation_row = annotation_row, cluster_rows = F)
annotation数据首先要组织成dataframe,dataframe中的rownames要和注释项进行对应,
而column就是要展示的注释条,每个column都会生成一个注释条来显示annotation_col/row 默认会有legend配合展示,annotation_legend = FALSE可以去掉legend
# Specify colors (自定义注释条颜色)ann_colors = list( Time = c("white", "firebrick"), CellType = c(CT1 = "#1B9E77", CT2 = "#D95F02"), GeneClass = c(Path1 = "#7570B3", Path2 = "#E7298A", Path3 = "#66A61E"))pheatmap(test, annotation_col = annotation_col, annotation_colors = ann_colors)pheatmap(test, annotation_col = annotation_col, annotation_row = annotation_row, annotation_colors = ann_colors)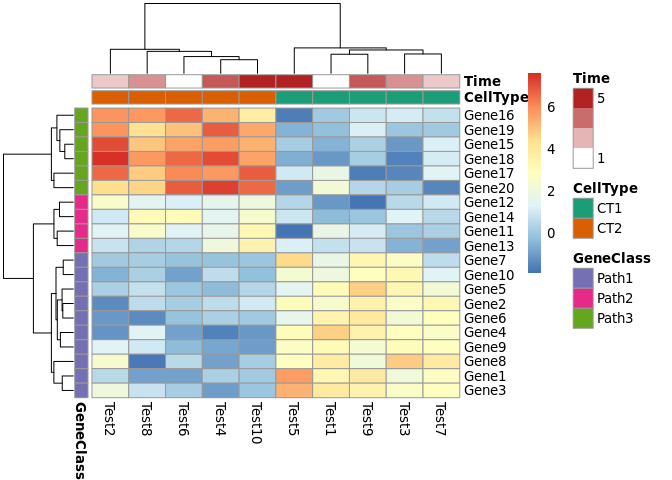
- 注释条颜色数据要组织成list(list的灵活性就在此处凸显出来了),
list中的每个元素名作为对应项(对应前述dataframe中的colnames),
同时可以进一步为attribute指定颜色,例如CT1 = "#1B9E77", CT2 = "#D95F02"
参考资料
pheatmap帮助文档